FAIRsharing Community Champions and in-house FAIRsharing staff have completed a massive curation effort begun more than 1 year ago, resulting in the curation and checking of every database-specific field across the entire FAIRsharing database registry. This resulted in more than thousands of edits over 2100 database records, and improves database transparency and readability across the entire research landscape. Access to these fields is available via the records themselves, our FAIRsharing Assistant and our Advanced Search. In this post we celebrate the success and hard work of our Community Champions, and let you know some of the ways in which this curation will be used in future. This work is similar to the efforts they made to enhance policy registry curation back in April, but is an order of magnitude larger. Please read on to learn more about this huge achievement.
The 17 databases-specific metadata fields were fully reviewed and updated, and range from data access and deposition through to sustainability, versioning and more. As a result, databases registered with FAIRsharing can now make their attributes more explicit and comparable by both humans and machines. FAIRsharing aligns with NIH efforts as well as multiple outputs of the RDA relating to data repository attributes. This means that creating and maintaining a database record with us means that your database automatically aligns with all of those community efforts. For more information, please see our documentation on how our database records align with community efforts.
Over the past year, eight of our Community Champions worked with our in-house team to review or annotate the more than 2100 records in our database registry.
From our Alumni:
- Enrique Wulff
- Geneviève Michaud
From our active Champions:
- Yojana Gadiya
- Lindsey Anderson
- Yuhe Liang
- Gabriel Pelletier
- Kyle Copas
- Kay Burrows
- Geta Mitrea
- Christian Bonatto Minella
- Our internal curation team: Delphine Dauga, Ramon Granell, Lea Girard, and Allyson Lister.
Their work is extraordinary, and makes evaluation and analysis of the research resource landscape more rewarding and easier than ever.
What are you interested in discovering? How many European repositories have open access? What about how many databases around the world incorporate versioning, or provide contact information for the data owners? You can filter and browse this database metadata via our FAIRsharing Assistant and our Advanced Search.
To whet your appetite, here are some statistics to showcase just a few of the characteristics of the databases in our registry. Want to include your database in our registry, or claim ownership of your database that is already registered? We have lots of documentation to help you get started adding new content.
For all of the following charts, you can find out more about the fields in FAIRsharing in our documentation in the Database Attributes and Conditions page.
85% of the databases in FAIRsharing have open access; likely there is some level of confirmation bias at play, as databases are much more likely to register with a service such as FAIRsharing if they are at least partially open. However, it is an impressive statistic nonetheless. Deposition is a more nuanced field, as knowledgebases (according to our definition) will always have a ‘not applicable’ value. Institutional repositories will generally have ‘controlled’ deposition restricted to institutional staff and collaborators only. However, even with these restrictions one third of registered databases have open deposition.
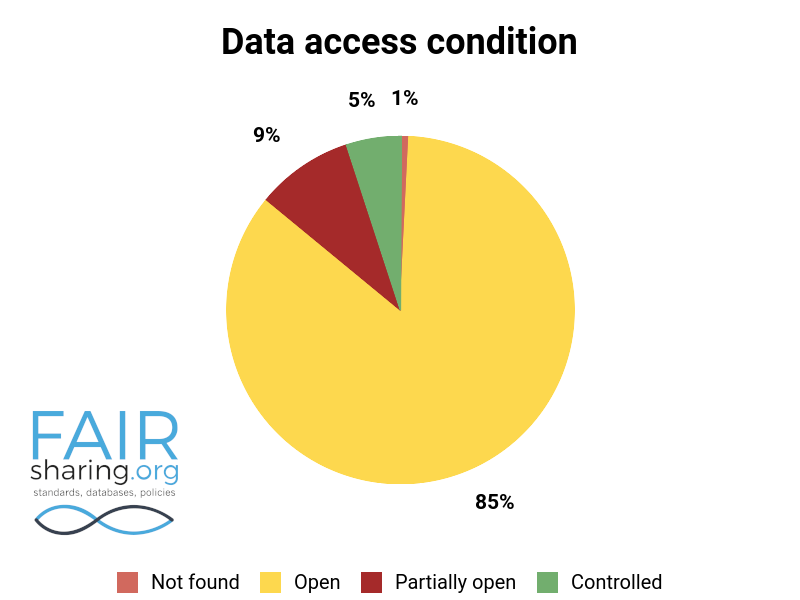
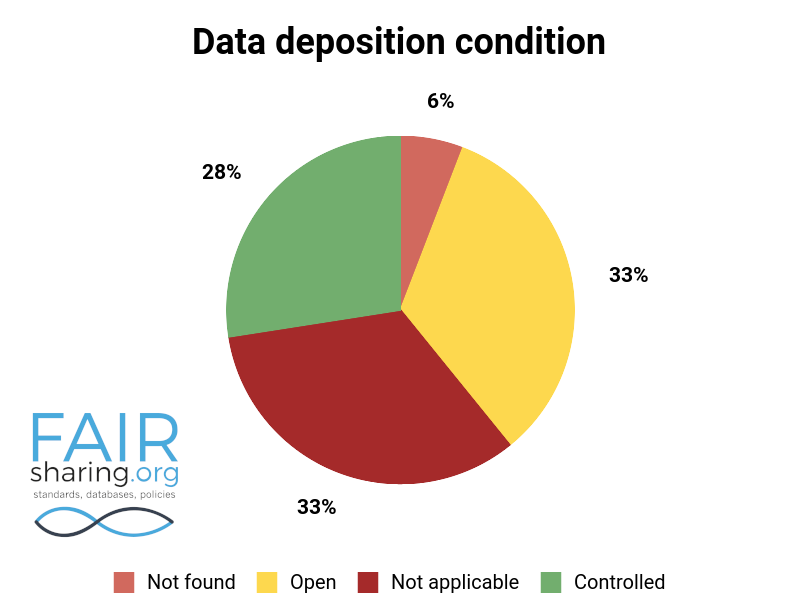
Additional fields include types of data curation, versioning, sustainability, preservation, availability for pre-publication review, citation to related publications, and contact information for data authors. You can see the distribution of these fields below.
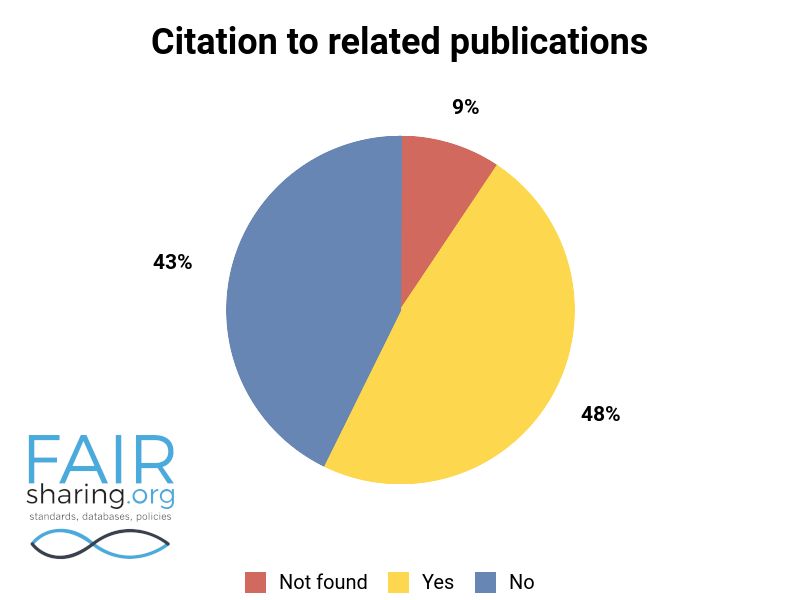
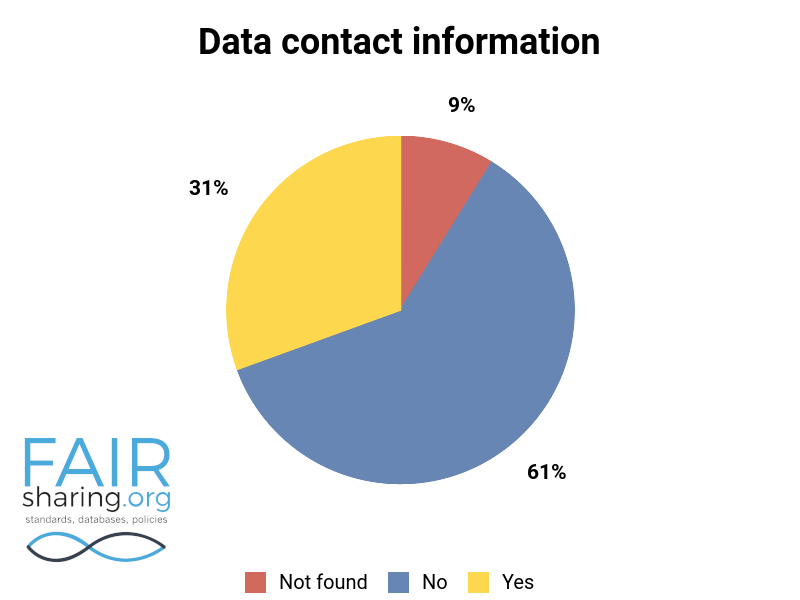
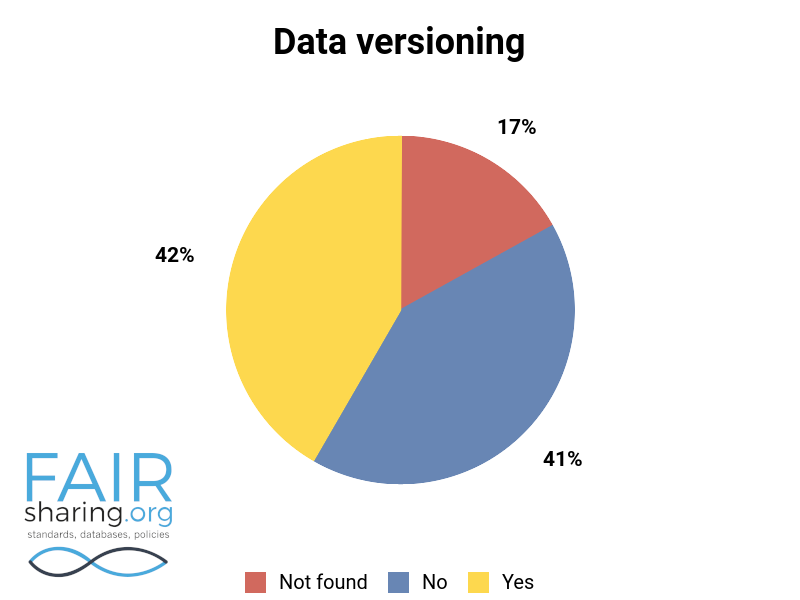
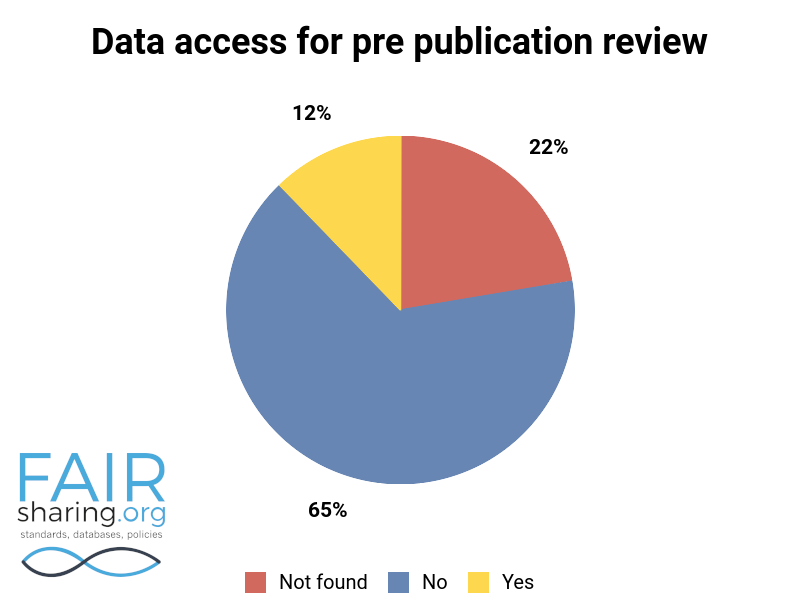
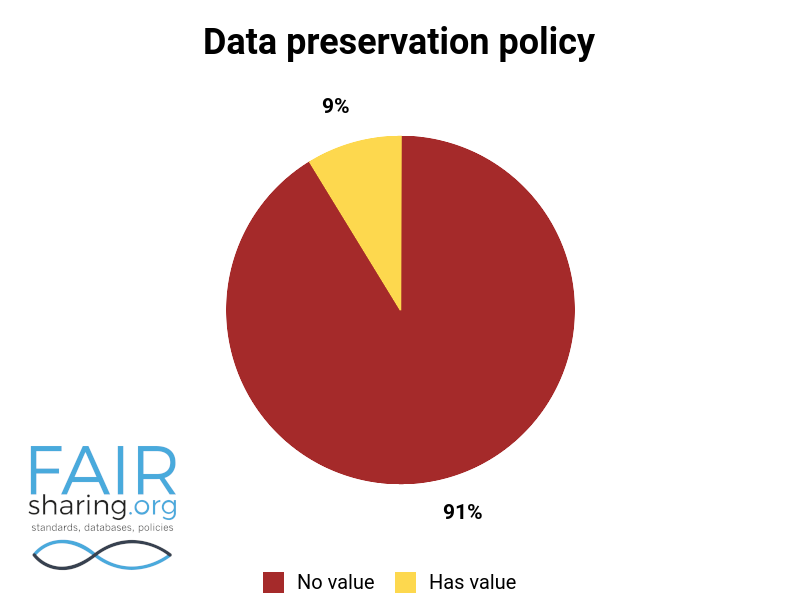
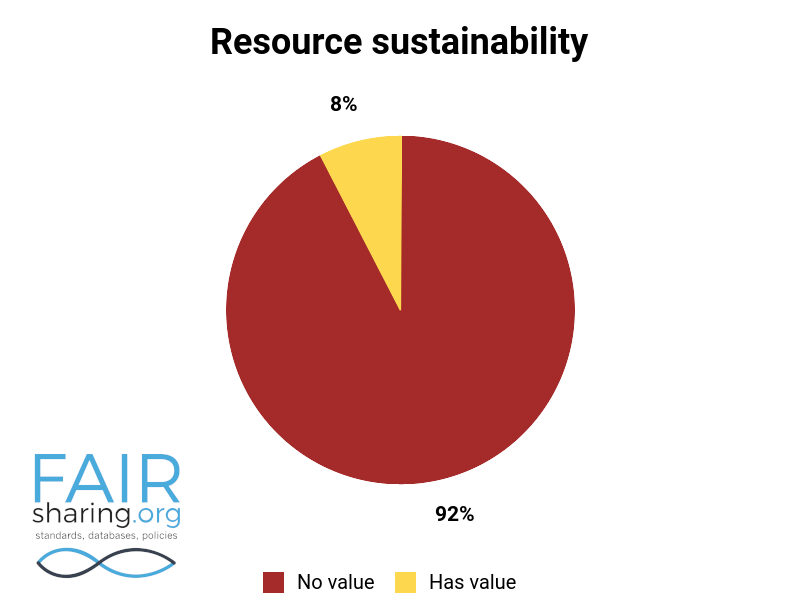
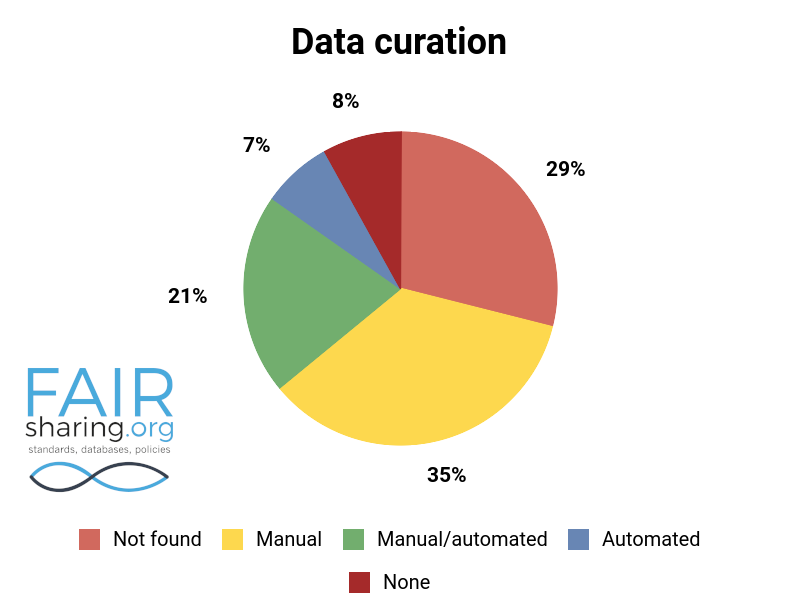
We are very thankful to all of our Champions, and especially to those who have contributed to this superlative effort within the database registry.
For more information about how different stakeholders can make use of FAIRsharing, please see our educational pages.
